Software Packages
More code/packages listed on our GitHub page
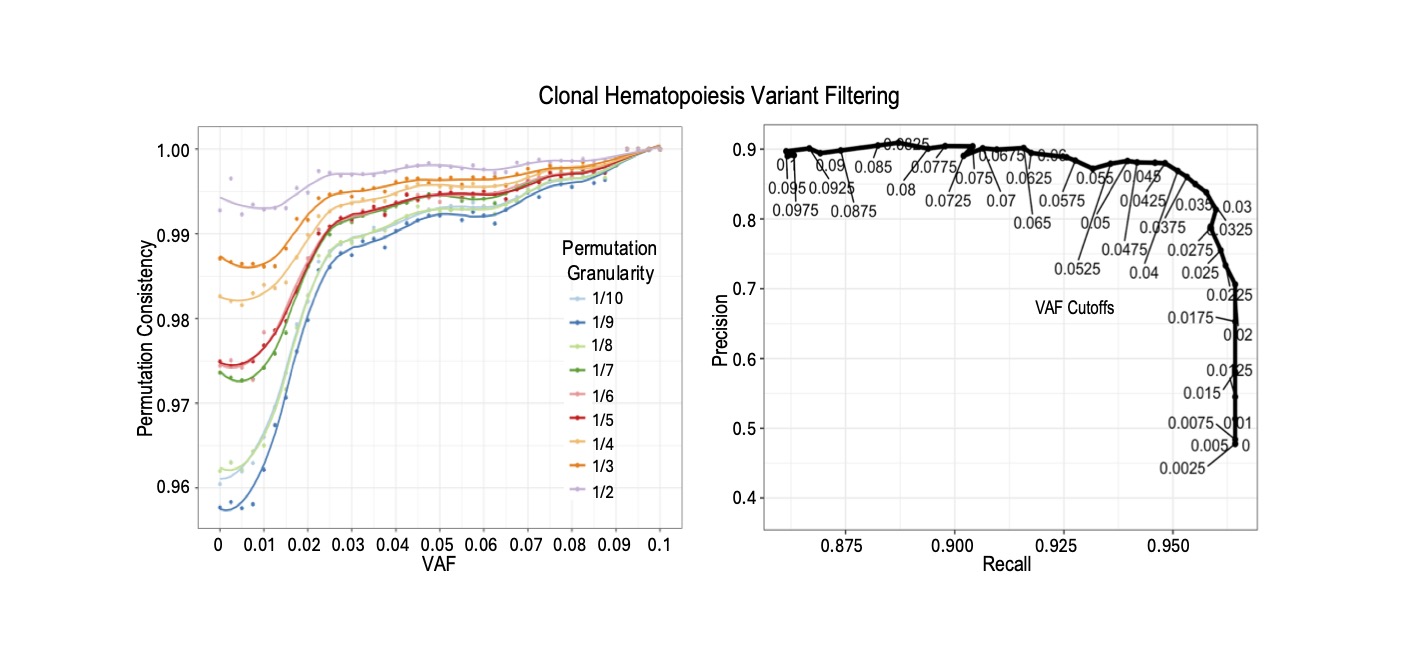
An R/GitHub package for estimating quality parameters and filtering clonal hematopoiesis variants from WES- or targeted-DNAseq-based mutation calls. The tool first uses permutation test to determine the optimal parameter values for a given dataset in CHIP filtering, e.g., VAF cutoffs. It then implements CH filtering using four groups of data metrics. We demonstrated its precision and recall by applying it to sequencing data cohorts across multiple solid cancer types in the ORIEN Network.
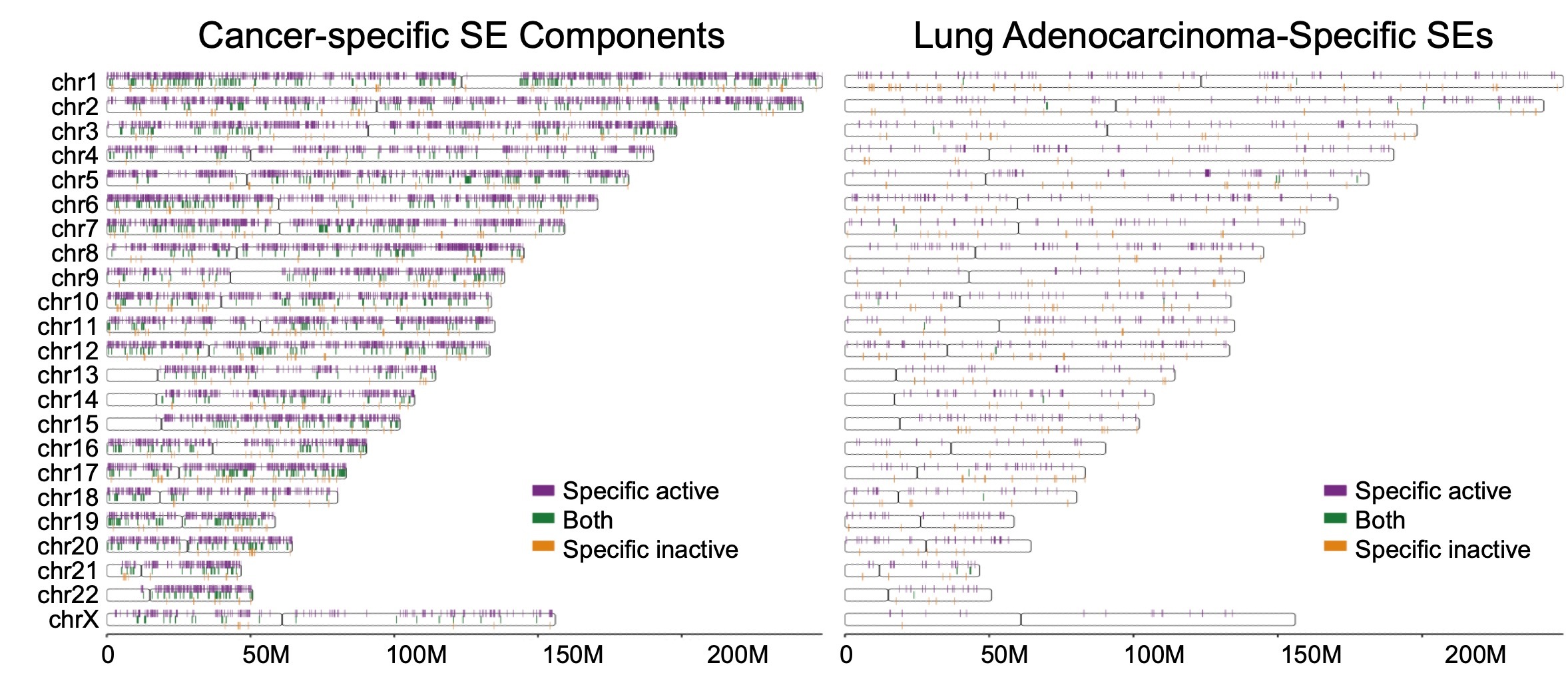
An R/GitHub package documenting high-resolution differences of super enhancer (SE) signatures across cancers. The sensitivity and specificity of cancer-specific SE identification are boosted by using mixture models to evaluate SE consensuses across 60 cell lines from 28 cancer types. Both cancer-specific-active and -inactive components of SEs are profiled here, enabling a pan-cancer systematic overview. Visualizing interface is built in the package to facilitate exploration.
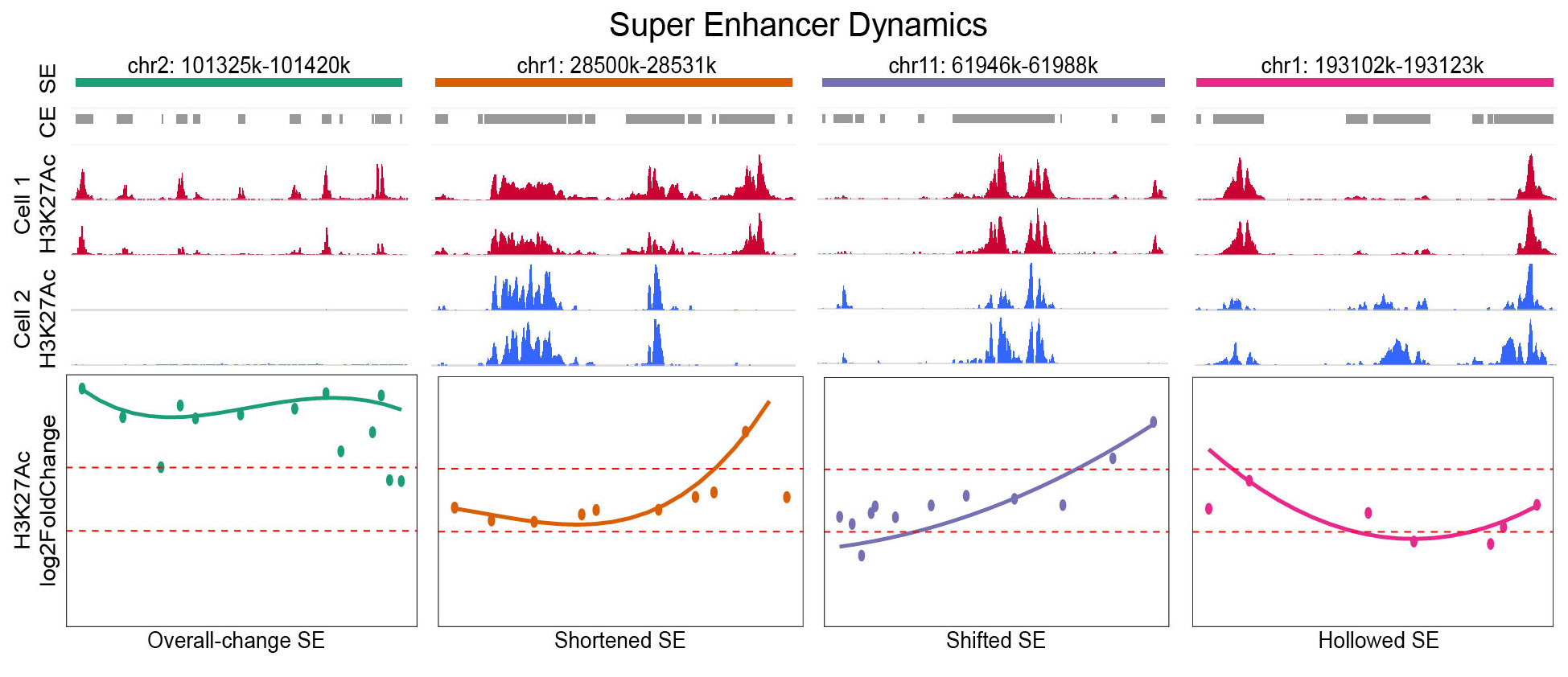
An R/GitHub package for detecting constitutive alterations of super enhancers (SE) using ChIP-seq. Four novel types of constitutive alterations as well as overall activity alterations are reported by this tool. Constitutive alterations usually demonstrate distinct functional characteristics in the way they regulate target genes. This tool elevates the sensitivity in detecting SE profile changes. It is highly recommended when SE profiles of similar cell conditions are compared.
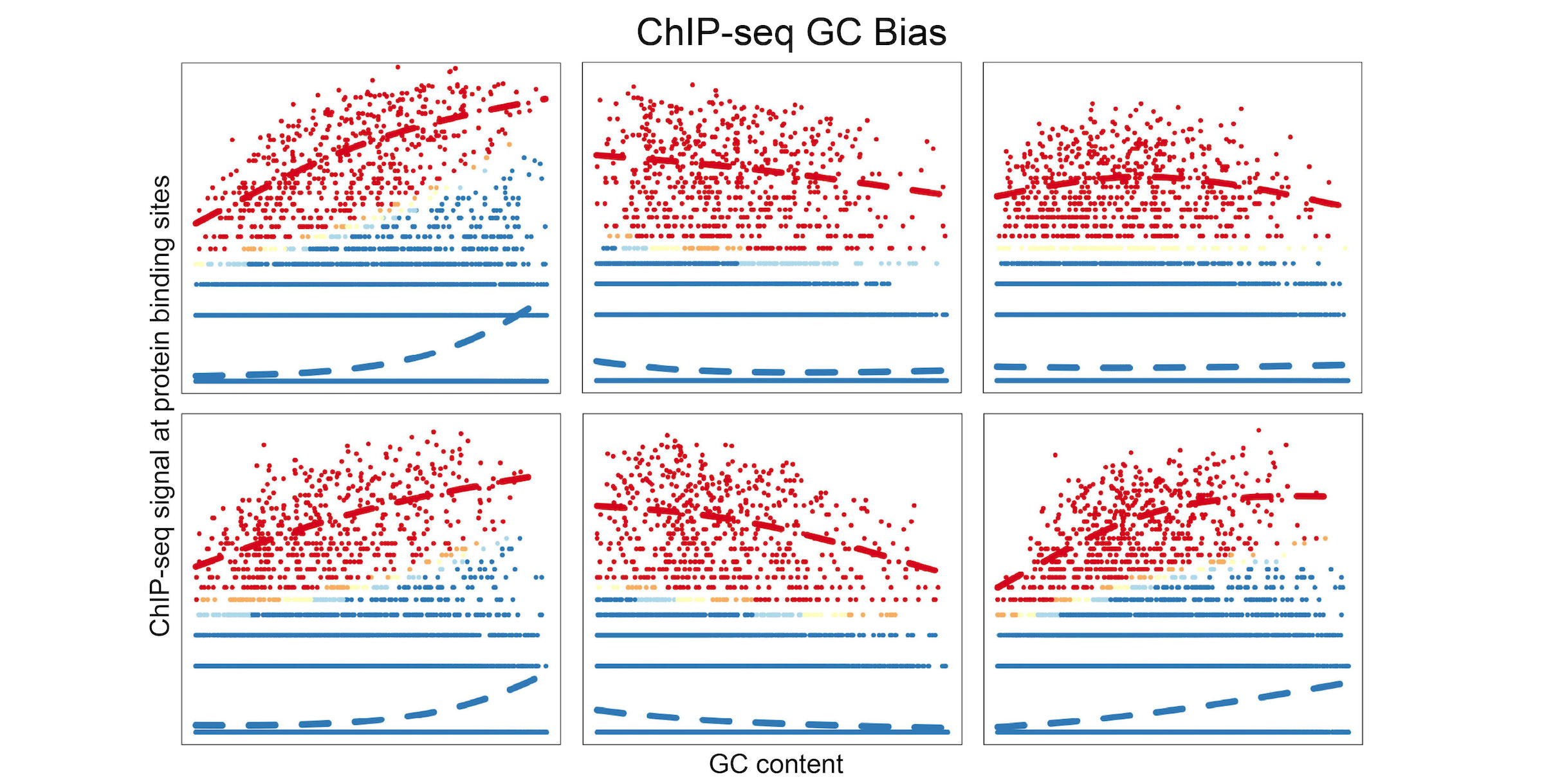
An R/Bioconductor package for ChIP-seq peak calling with adjustment of GC-content bias. GC-content bias is firstly estimated with generalized linear mixture models using effective GC strategy, then applied to the estimation of peak enrichment. This package also provides alternative option to correct GC-content bias for a given list of genomic regions across multiple samples. Package is also available on GitHub.
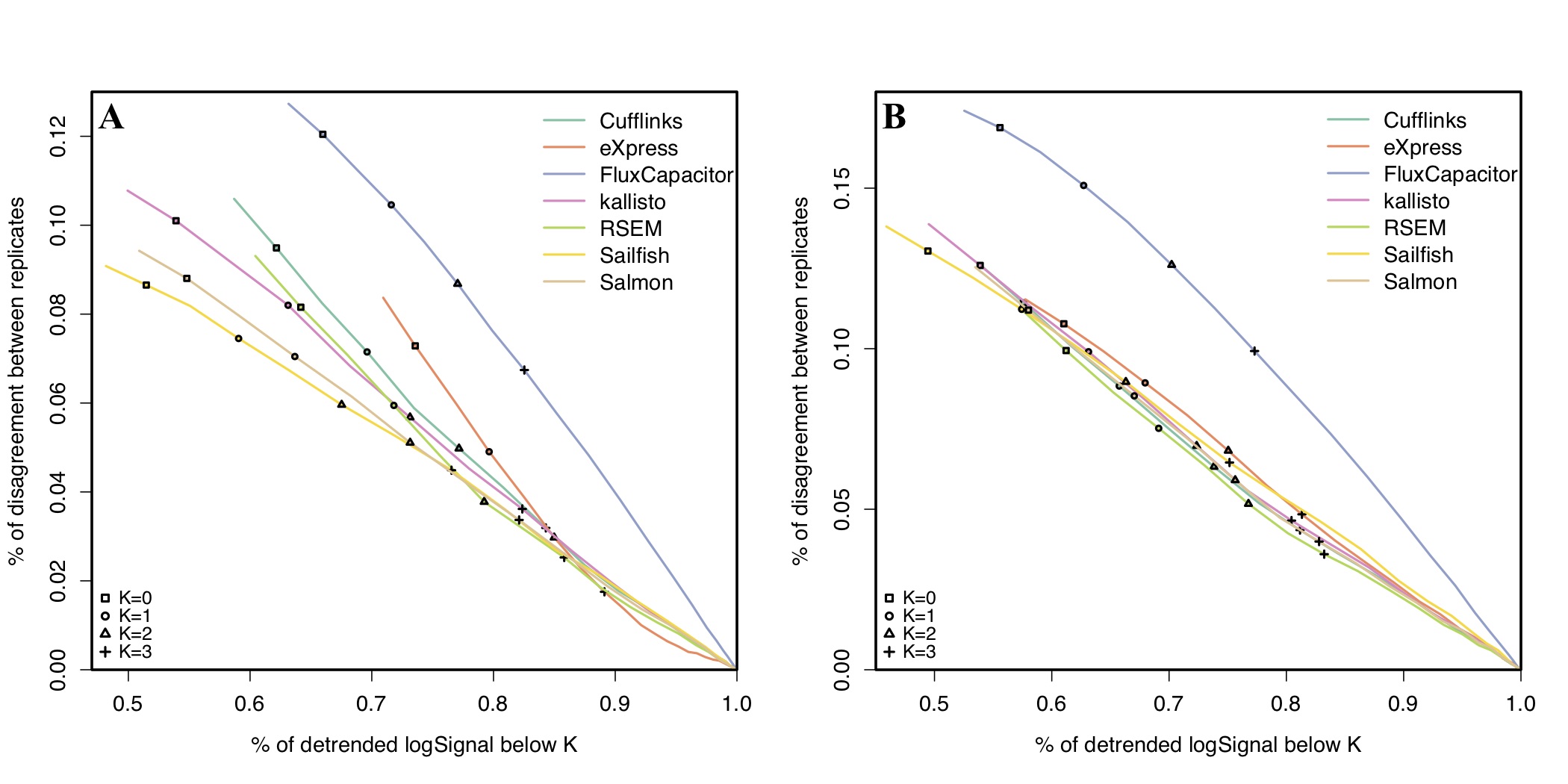
An R/Bioconductor package providing evaluation and visualization of several quantitative benchmarks for RNA-seq quantification pipelines, accompanied with an easy-to-use web tool. Quantifications of genes, transcripts, junctions or exons in two compared conditions by each pipeline with necessary meta information should be organized into numeric matrices in order to proceed with the evaluation. Package is also available on GitHub.
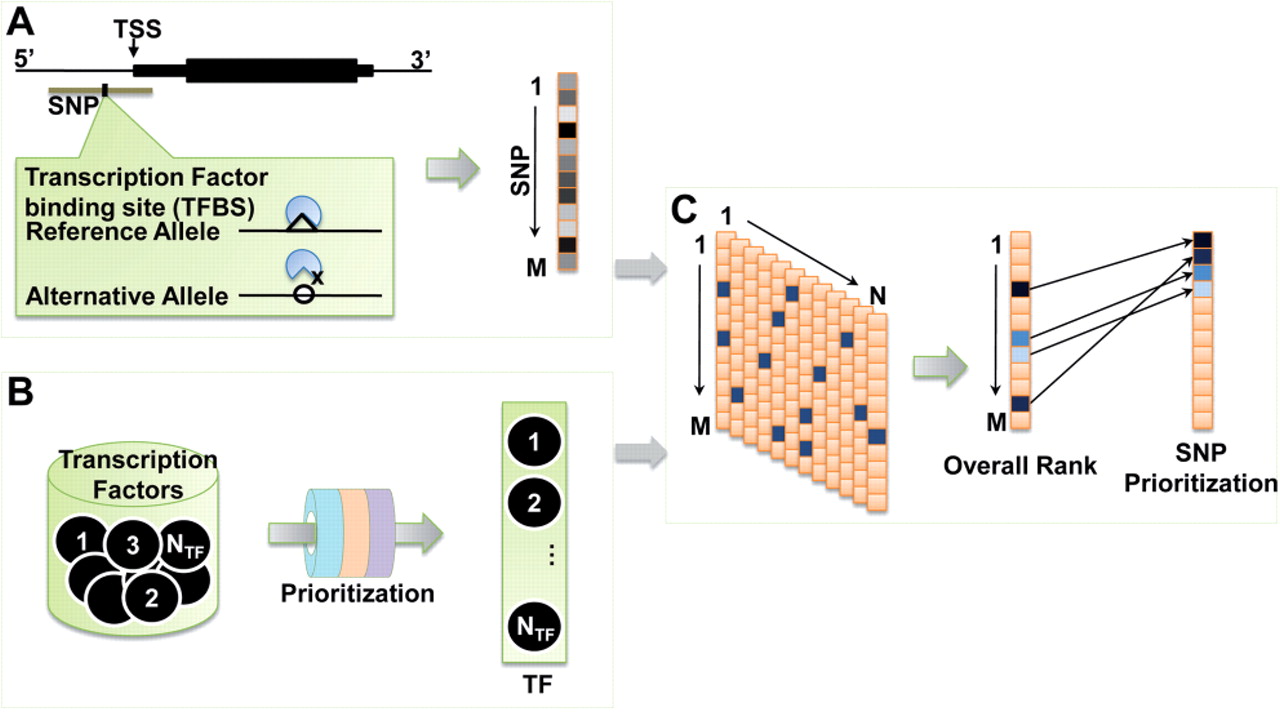
An R-based algorithm that integrates multiple bioinformatics tools/databases for prioritizing regulatory SNPs, i.e., the SNPs in the promoter regions that potentially affect phenotype through transcription disruption. regSNPs considers degenerative features of transcription factor (TF) binding motifs. It calculates SNP-caused alterations of TF binding and integrates that with the phenotypic effects of the altered TFs.